 |
|
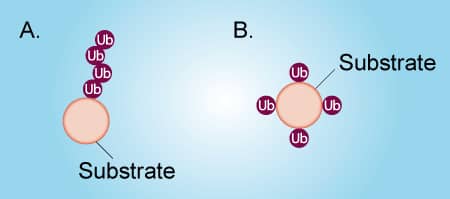
Figure 1. Poly-ubiquitination vs Multi-mono-ubiquitination |
Ubiquitin can be attached to a protein substrate via two distinct mechanisms (Figure 1). Poly-ubiquitination occurs when Ubiquitin molecules are attached end-to-end to a single lysine residue on a substrate protein to form a poly-ubiquitin chain (Figure 1A). Alternatively, multi-mono-ubiquitination is the attachment of a single Ubiquitin molecule to multiple lysine residues on a substrate protein (Figure 1B). It is important to distinguish between poly-ubiquitination and multi-mono-ubiquitination because these different types of ubiquitination lead to different functions of the substrate protein. To complicate matters, a poly-ubiquitinated protein and a multi-mono-ubiquitinated protein look very similar by SDS-PAGE and Western blot. Fortunately, it is relatively simple to differentiate between poly-ubiquitination and multi-mono-ubiquitination by performing in vitro ubiquitin conjugation reactions. The protocol below describes in detail how to determine if your protein of interest is poly-ubiquitinated or multi-mono-ubiquitinated. Click on the links below to view listings of Boston Biochem® proteins, enzymes, and buffers.
Materials and reagents:
| Material or Reagent | Stock Concentration |
| E1 Enzyme | 5 µM |
| E2 Enzymei | 25 µM |
| E3 Ligaseii | 10 µM |
| 10X E3 Ligase Reaction Buffer | 10X - (500 mM HEPES, pH 8.0, 500 mM NaCl, 10 mM TCEP) |
| Ubiquitin | 1.17 mM (10 mg/mL) |
| Ubiquitin No K | 1.17 mM (10 mg/mL) |
| MgATP Solution | 100 mM |
| SDS-PAGE sample buffer – if not using reaction products for downstream applications | 2X |
| EDTA or DTT – if using reaction products for downstream applications | 500 mM (EDTA);1 M (DTT) |
| Microcentrifuge tubes | |
| Water Bath (37 °C) | |
| Western Blot Equipment |
iEach E2 enzyme functions with only a subset of E3 ligases and some E3’s are more promiscuous than others.
iiThe E3 ligase will likely need to be supplied by the user, but we do offer a small selection.
Procedure for distinguishing between poly-ubiquitination and multi-mono-ubiquitination
 |
|
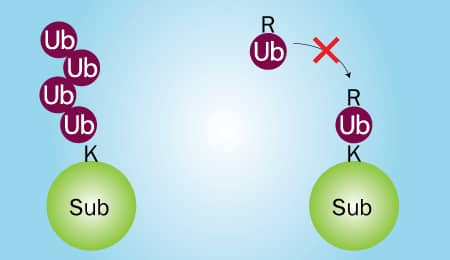
Figure 2. Wild Type Ubiquitin vs Ubiquitin No K |
Two in vitro Ubiquitin conjugation reactions will need to be performed: 1) one that contains wild type Ubiquitin and 2) one that contains Ubiquitin No K, a mutant in which all 7 lysines have been mutated to arginines. Wild type Ubiquitin can be conjugated to substrate proteins and is capable of forming chains (Figure 2; left). Ubiquitin No K can also be conjugated to substrate proteins, but it is unable to form chains due to its lack of lysine residues (Figure 2; right). Therefore, if your substrate is poly-ubiquitinated, high molecular weight bands will be generated in reaction 1, but not reaction 2 (Figure 3A). Conversely, if your substrate is multi-mono-ubiquitinated, high molecular weight bands will be generated in both reaction 1 and reaction 2 (Figure 3B). Note that reaction 1 will look the same for both poly-ubiquitinated and multi-mono-ubiquitinated substrates.
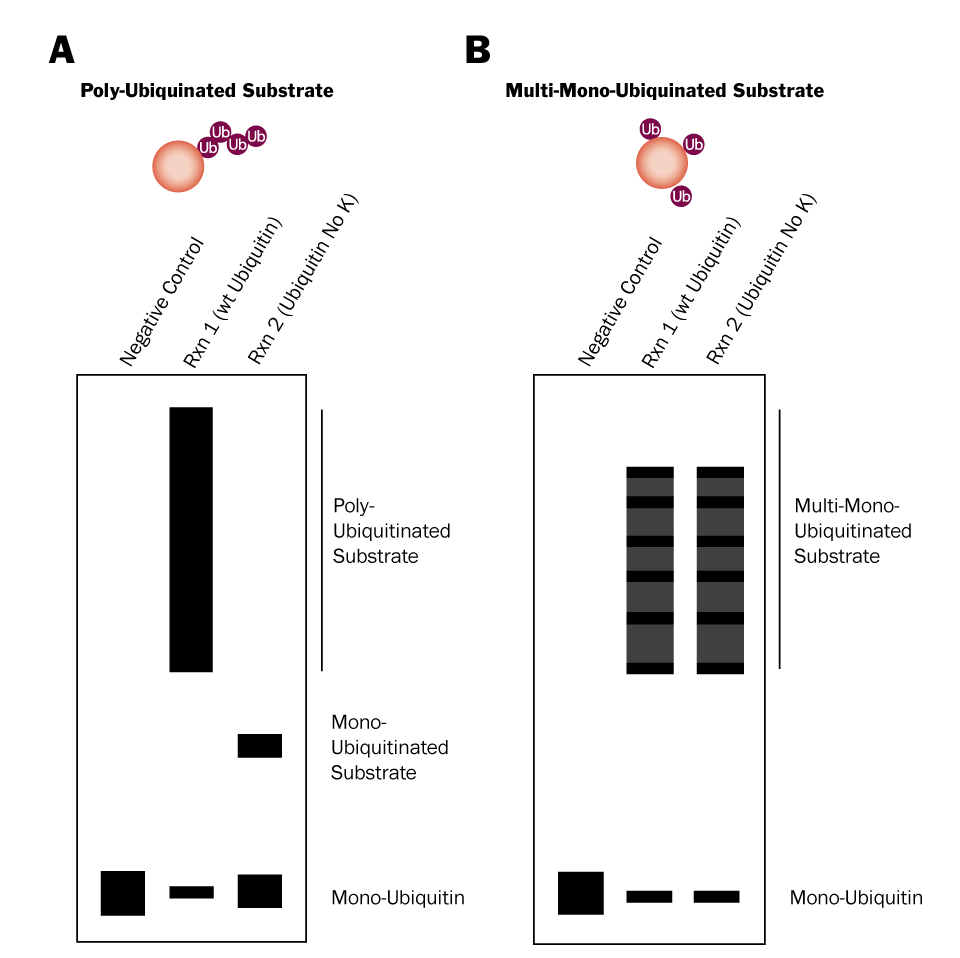 Figure 3. Differentiation Between Poly-Ubiquitination and Multi-Mono-Ubiquitination Using Ubiquitin No K |
Procedure for 25 µL reactions (scale as needed):
- For reaction 1, combine the indicated volume of each component listed in the table below, in the order shown, in a microcentrifuge tube. For a negative control reaction replace the MgATP Solution with dH2O.
Reaction 1
| Reagent | Volume | Working Concentration |
| dH2O | X µL (to 25 µL; dependent on volume of substrate and E3 ligase) | N/A |
| 10X E3 Ligase Reaction Buffer | 2.5 µL | 1X - (50 mM HEPES, pH 8.0, 50 mM NaCl, 1 mM TCEP) |
| Ubiquitin | 1 µL | Approximately 100 µM |
| MgATP Solution | 2.5 µL | 10 mM |
| Substrate | X µLiii | 5-10 µM |
| E1 Enzyme | 0.5 µL | 100 nM |
| E2 Enzyme | 1 µL | 1 µM |
| E3 Ligase | X µLiv | 1 µM |
iiiThe volume needed will depend on the stock concentration of your substrate.
ivThe volume needed will depend on the stock concentration of your E3 ligase.
- For reaction 2, combine the indicated volume of each component listed in the table below, in the order shown, in a microcentrifuge tube. The only difference between reaction 1 and reaction 2 is the substitution of Ubiquitin No K for wild type Ubiquitin.
Reaction 2
| Reagent | Volume | Working Concentration |
| dH2O | X µL (to 25 µL; dependent on volume of substrate and E3 ligase) | N/A |
| 10X E3 Ligase Reaction Buffer | 2.5 µL | 1X - (50 mM HEPES, pH 8.0, 50 mM NaCl, 1 mM TCEP) |
| Ubiquitin No K | 1 µL | Approximately 100 µM |
| MgATP Solution | 2.5 µL | 10 mM |
| Substrate | X µLiii | 5-10 µM |
| E1 Enzyme | 0.5 µL | 100 nM |
| E2 Enzyme | 1 µL | 1 µM |
| E3 Ligase | X µLiv | 1 µM |
iiiThe volume needed will depend on the stock concentration of your substrate.
ivThe volume needed will depend on the stock concentration of your E3 ligase.
- Incubate reaction 1 and reaction 2 in a 37 °C water bath for 30-60 minutes.
- Terminate the reactions. See the table below for the appropriate method of termination.
| Are you using reaction products for downstream enzymatic applications? | Termination method | Volume (final concentration) |
| No | SDS-PAGE sample buffer | 25 µL (1X) |
| Yes | EDTA or DTTv | 0.5 µL EDTA (20 mM) or 1 µL DTT (100 mM) |
vEDTA and DTT are equally effective at terminating the reaction; determining which to use will depend on the intended downstream enzymatic application of the reaction products.
- Analyze Ubiquitin conjugation reactions.
- Separate the reaction products by SDS-PAGE and transfer them to a PVDF or nitrocellulose membrane.
- Perform a Western blot using an anti-Ubiquitin antibody.
- Compare your Western blot to figure 3 above to determine if your substrate is poly-ubiquitinated or multi-mono-ubiquitinated.*
*It is possible for a substrate to be both poly-ubiquitinated and multi-mono-ubiquitinated. If this is the case the Ubiquitin No K will still prevent poly-ubiquitination and the highest molecular weight protein species should disappear from reaction 2. Contact us for technical assistance!